Note
Click here to download the full example code
Plot sulci on the flatmap¶
The sulci are defined in a sub-layer of the sulci layer in <filestore>/<subject>/overlays.svg.
The parameter with_sulci in quickflat.make_figure controls displaying the sulci on the surface.
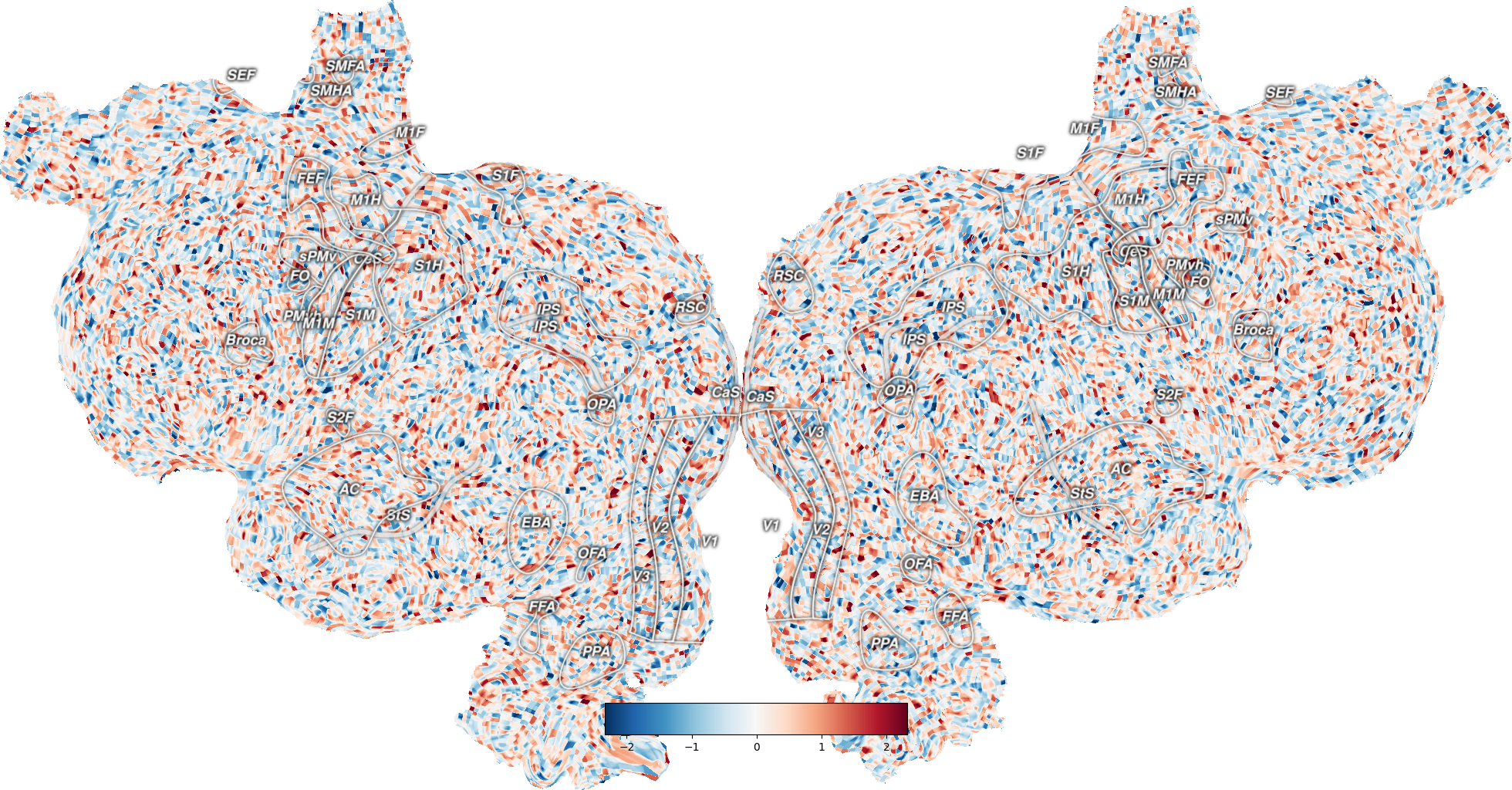
Out:
Generating curvature surface info...
/opt/hostedtoolcache/Python/3.9.12/x64/lib/python3.9/site-packages/scipy/sparse/linalg/_dsolve/linsolve.py:322: SparseEfficiencyWarning: splu requires CSC matrix format
warn('splu requires CSC matrix format', SparseEfficiencyWarning)
Failed to get connection
** (inkscape:5209): CRITICAL **: 01:27:48.014: dbus_g_proxy_new_for_name: assertion 'connection != NULL' failed
** (inkscape:5209): CRITICAL **: 01:27:48.014: dbus_g_proxy_call: assertion 'DBUS_IS_G_PROXY (proxy)' failed
** (inkscape:5209): CRITICAL **: 01:27:48.014: dbus_g_connection_register_g_object: assertion 'connection != NULL' failed
Failed to get connection
** (inkscape:5213): CRITICAL **: 01:27:48.884: dbus_g_proxy_new_for_name: assertion 'connection != NULL' failed
** (inkscape:5213): CRITICAL **: 01:27:48.884: dbus_g_proxy_call: assertion 'DBUS_IS_G_PROXY (proxy)' failed
** (inkscape:5213): CRITICAL **: 01:27:48.884: dbus_g_connection_register_g_object: assertion 'connection != NULL' failed
import cortex
import numpy as np
np.random.seed(1234)
# Create a random pycortex Volume
volume = cortex.Volume.random(subject='S1', xfmname='fullhead')
# Plot a flatmap with the data projected onto the surface
# Highlight the curvature and display the sulci
_ = cortex.quickflat.make_figure(volume,
with_curvature=True,
with_sulci=True)
Total running time of the script: ( 0 minutes 9.313 seconds)